This script parses out unaligned columns.
E.g. your data is formatted in the following:
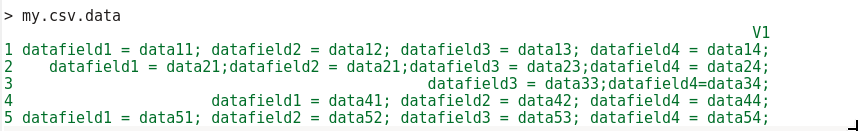
Create your strings as:

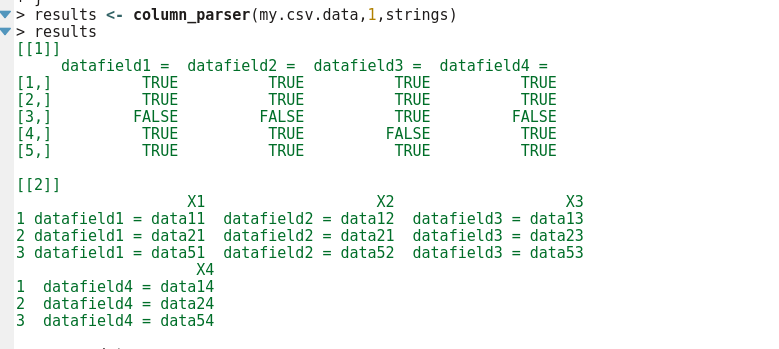
And output:
# This script parses unaligned columns
# It takes three arguments
# 1) The Dataset name
# 2) The column number you want to parse == 1 (The original need required this to be inputted. Now you should only input 1 column that you want to get parsed and pass this as 1)
# 3) A character vector called "strings" that contains the fields you want to extract
# It will *only* return those content fields where all of the 'strings' requirements are met.
# - you will not get NA-values
# However, the output is a list of 2. list[[1]] is the TRUE/FALSE table so you can see which rows contain or do not
# contain the strings you wanted to get out
# The returned list second element list[[2]] is the dataframe of parsed content fields, no NA values. All FALSE rows from
# the TRUE/FALSE table are not in the second list.
# There is no error handling. IF your content field contains a delimiter in the column you want to extract, the code will parse it
# and your data will look wrong, but you should receive a generic WARNING/ERROR from R anyway
# The parser seems to give some warnings/errors when you have matching parseable fields, e.g. "dataX = " vs. "dataX = ", a decent workaround
# is to gsub the field and change that data to something else and re-run. This has been found to work.
# Do not trust the results with generic R warnings/errors, GSUB your data and re-run
column_parser <- function(dataset, content_field, strings){
returnlist <- list()
content <- dataset[,content_field]
content <- strsplit(content, ";") # You can specify any delimiter here
content <- lapply(content,trimws)
content_truefalse <- sapply(strings, function(x) grepl(x, content))
investigate_these <- dataset[rowSums(content_truefalse)>length(strings)-1,]
returnlist[[1]] <- content_truefalse
investigate_these <- strsplit(investigate_these,';')
inv1 <- lapply(investigate_these,function(x) grep(paste(strings,collapse="|"),x,value=TRUE))
inv2 <- data.frame(matrix(unlist(inv1), nrow=length(inv1), byrow=T),stringsAsFactors=FALSE)
returnlist[[2]] <- inv2
return(returnlist)
}